CoMoFinder
-- MANUAL --
Description of CoMoFinder
CoMoFinder strives to discover reliable composite network motifs in co-regulatory networks which consist of microRNAs, transcriptional regulators and genes.
We provide two versions of the command line interface of CoMoFinder to satisfy users with different needs. Both versions are distributed in Java archive files (JARs). The first version, i.e. CoMoFinder.jar, is an integrated one which executes the algorithm plainly and generates all the randomized batch files and final statistics of the composite subgraphs to the specialized directories. The second version (including referenceCount.jar, batchProcess.jar and significance.jar) is a separated one which can be executed on computing clusters. Therefore for users who want to run CoMoFinder on their own desktops or laptops, we recommend them to use the integrated version, while for users who can submit their jobs to computing clusters we encourage them to use the separated version since the parallel process will significantly reduce the overall running time of the algorithm. In order to give more flexibility to users and save as much running time of the algorithm as possible, we separated the output process of the composite subgraphs (getSpecificMotifs.jar) from the main program. So users can specify the motifs of their interests and output the corresponding motifs to the files after the program discovers all the motifs from the input networks. Here we want to stress that during the output process all the motifs are written to the disks instantly instead of storing them into the memory first and writing them to the disk all at once, which means the memory usage of our algorithm is much more economic compared to FANMOD / WaRSwap when outputting the motifs. Also, we employed a Java Fork/Join framework in our algorithm that implements a work-stealing strategy to parallel the composite subgraph enumeration process [1, 2], which makes our algorithm several magnitudes faster than FANMOD / WaRSwap in multi-cores scenario.
Requirements of CoMoFinder
As CoMoFinder was developed in Java, we require users to have JDK 1.6 (or higher) installed on their operating systems. Also, a third-party jar fork/join framework is used in CoMoFinder, which has been included in our compressed file. Users can find it under the fold “lib/ jsr166y-1.7.0.jar”. Please keep all the CoMoFinder related jars and the “lib” folder under the same directory.
Usage of CoMoFinder
In this document, a dollar (“$”) sign at the start of a line in the examples represents the shell prompt of the operating system. There is no need to type it. A bracket (“[]”) sign means the parameters are optional. Assuming that you have the Java interpreter on the path of your operating system, to run the jar files is simply as follows:
$ java –jar XXX.jar [parameters]
List of parameters
==============================================================================
To run both versions of command line interfaces of CoMoFinder, several parameters need to be provided to the algorithm. There are no restrictions on the order of the parameters. The following is a full list of parameters needed for our algorithm:
-ms : specify the motif size for the algorithm, e.g. 4. The default value is 3.
-nr : specify the number of threads used in the algorithm, e.g. 4. By default, the program detects the available processors on the running computer and assigns subtasks to each of them.
-fp: specify the input file for the algorithm, e.g. “sample_input_network.txt”. This parameter must be provided.
-bfd: specify the output directory for the composite subgraphs count of the random networks ensemble, e.g. “output”. This parameter must be provided.
-ens: specify the size of the random network ensemble, e.g. 500. Default value is 1000.
-ite: specify the maximal number of iterations in the edge swap process within each randomization procedure, e.g. 50. Default value is 100.
-mfn: specify the final motif file name, e.g. “output/motif.txt”. This parameter must be provided.
-scn: specify the file name of the observed composite subgraphs count, e.g. “reference_count.txt”. This parameter must be provided.
-bn: specify the batch index for the current task, e.g 8. This parameter must be provided.
-list: specify the motifs that need to be output to the files, e.g. motifs_4.txt. This parameter must be provided.
-opd: specify the output directory for the detected motifs, e.g. output/subgraphList. This parameter must be provided.
==============================================================================
For integrated version CoMoFinder.jar:
A complete command line to run CoMoFinder.jar should be something like:
$ java –jar CoMoFinder.jar [-ms 3] [-nr 4] [-ens 100] –fp sample_input_network.txt –bfd output/batchFiles –mfn sig_motif.txt
For separated version:
A complete command line to run referenceCount.jar should be something like:
$ java –jar referenceCount.jar [-ms 3] [-nr 4] –fp sample_input_network.txt –scn output/ref_count.txt
A complete command line to run batchProcess.jar should be something like:
$ java –jar batchProcess.jar [-ms 3] [-nr 4] –fp sample_input_network.txt –bfd output/batchFiles –bn 4
A complete command line to run significance.jar should be something like:
$ java –jar significance.jar –bfd output/batchFiles –scn output/ref_count.txt –mfn sig_motif.txt
To output the motifs of interests to disks, a complete command line to run getSpecificMotifs.jar should be something like:
$ java –jar getSpecificMotifs.jar –list motifs.txt –fp sample_input_network.txt –opd output/subgraphList
Format of the input file
The input file format for the parameter “fp” should contain one line for each edge in your network, each line must contain at least four integers. It may contain up to five integers, only if for multiples types of edges (e.g. when PPIs are integrated into the network the users can specify another edge type for the PPIs). By default, the edge type for each edge will be set to 1. A line in the input file looks like this:
sourceIDtargetID sourceType targetType [edgeType]
, where the separation between each element should be a tab-delimited (“\t”) sign. Since our main focus is co-regulatory networks, the line is automatically interpreted as a directed edge from sourceID to targetID. The sourceType and targetType designate the node types of the source and target, i.e. 0 (microRNAs), 1 (transcriptional regulators) or 2(genes) in our algorithm. Please bear in mind that all node IDs need to be consecutive integers and the node IDs of miRNAs should always be smaller than those of transcriptional factors and genes. A sample input network “sample_input_network.txt” could be found in the compressed file.
The input file format for the parameter “list” is simply a single column that composes of the composite motif labels found by the program. Each line contains one motif label. A simple illustration is as follows:
001101000_012
011001100_012
…
Interpretation of the output
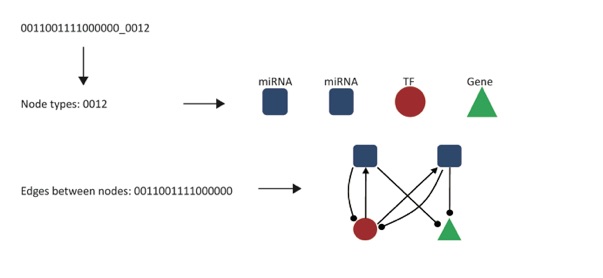
Our algorithm outputs all the information for each composite subgraph observed in the given network, including the “Subgraph_Label”, “occupancy”, “mean_frequency”, “standard_deviation”, “ZScore”, “pValue” and “frequency_in_real_network”. All the definitions and meanings for each measurement could be found in the main text. Users can define their own cutoffs and select corresponding composite subgraphs as motifs. Here we give an example on how to convert the Subgraph_Label to the composite subgraphs. A subgraph label consists of two parts: edges between nodes and node types, which are concatenated by the “_” sign. Assume the subgraph label is “0011001111000000_0012”, then “0011001111000000” stands for the edges between nodes, and “0012” are the node types for the four nodes. So from the node types part users can find that the first two nodes are microRNAs, the third node is a TF and the last node is a gene. From the edges part users can find that there are two regulations from the first microRNA to the TF and gene, two regulations from the second microRNA to the TF and gene, and two regulations from the TF to both of the microRNAs. An illustration is as follows:
1.Holmes, D. and D. Lea, Introduction to concurrent object-oriented programming in Java. Technology of Object-Oriented Languages - Tools 33, Proceedings, 2000: p. 457-457.
2.Zambas, C. and M. Lujan, Introducing aspects to the implementation of a Java Fork/Join framework. Algorithms and Architectures for Parallel Processing, Proceedings, 2008. 5022: p. 294-304.